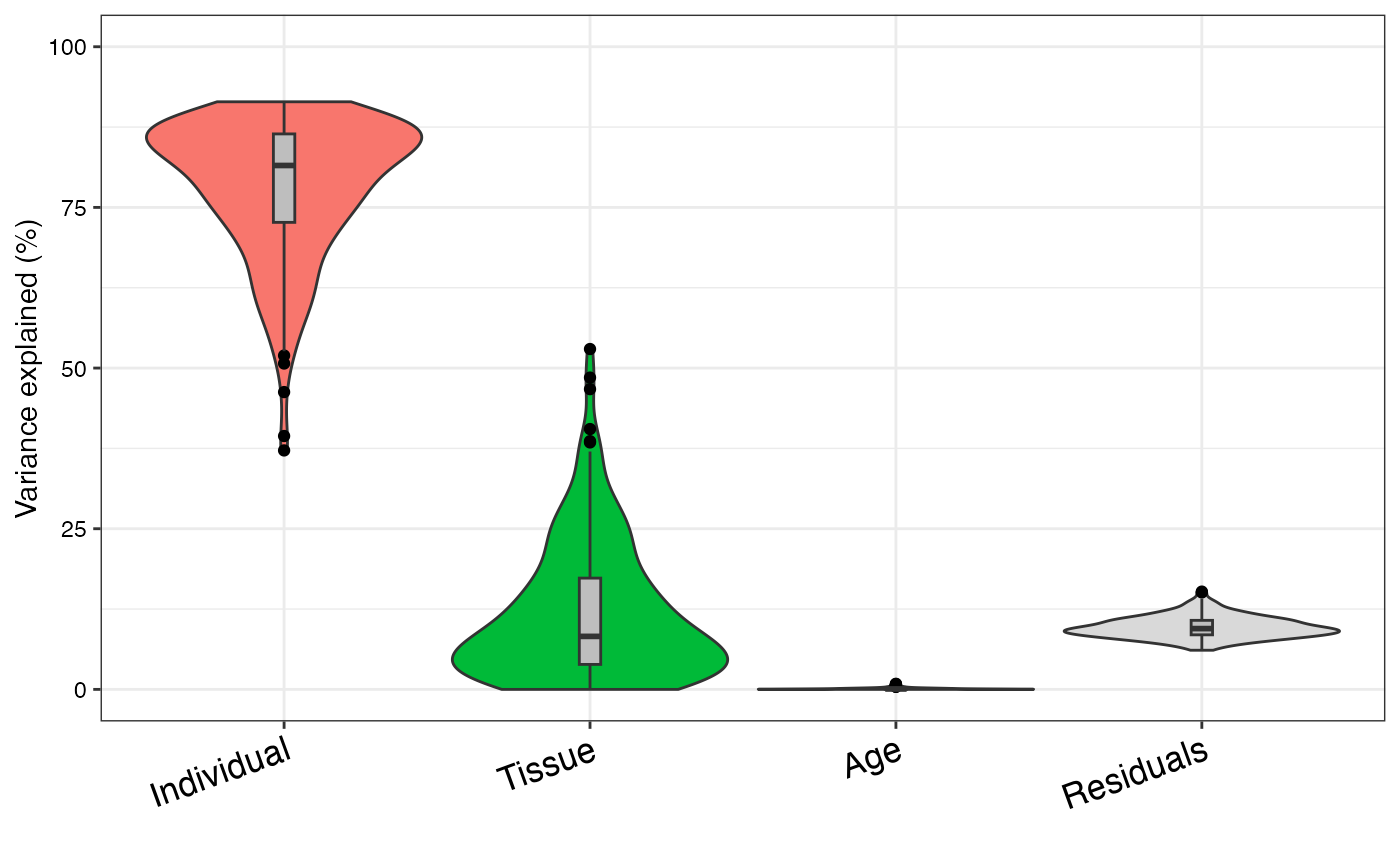
Violin plot of variance fraction for each gene and each variable
Usage
plotVarPart(
obj,
col = c(ggColorHue(ncol(obj) - 1), "grey85"),
label.angle = 20,
main = "",
ylab = "",
convertToPercent = TRUE,
...
)
# S4 method for class 'matrix'
plotVarPart(
obj,
col = c(ggColorHue(ncol(obj) - 1), "grey85"),
label.angle = 20,
main = "",
ylab = "",
convertToPercent = TRUE,
...
)
# S4 method for class 'data.frame'
plotVarPart(
obj,
col = c(ggColorHue(ncol(obj) - 1), "grey85"),
label.angle = 20,
main = "",
ylab = "",
convertToPercent = TRUE,
...
)
# S4 method for class 'varPartResults'
plotVarPart(
obj,
col = c(ggColorHue(ncol(obj) - 1), "grey85"),
label.angle = 20,
main = "",
ylab = "",
convertToPercent = TRUE,
...
)Value
Makes violin plots of variance components model. This function uses the graphics interface from ggplot2. Warnings produced by this function usually ggplot2 warning that the window is too small.
Examples
# load library
# library(variancePartition)
library(BiocParallel)
# load simulated data:
# geneExpr: matrix of gene expression values
# info: information/metadata about each sample
data(varPartData)
# Specify variables to consider
# Age is continuous so we model it as a fixed effect
# Individual and Tissue are both categorical, so we model them as random effects
form <- ~ Age + (1 | Individual) + (1 | Tissue)
varPart <- fitExtractVarPartModel(geneExpr, form, info)
# violin plot of contribution of each variable to total variance
plotVarPart(sortCols(varPart))