Run hierarchical clustering preserving sequential order of entries
Arguments
- X
data matrix were *rows* are features in sequential order
- gr
GenomicRanges object corresponding to rows in X
- alpha
mixture parameter weigting correlations (alpha=0) versus chromosome distances (alpha=1)
Details
Use hclustgeo in ClustGeo package to generate hierarchical clustering that preserves sequential order.
Chavent, et al. 2017. ClustGeo: an R package for hierarchical clustering with spatial constraints. arXiv:1707.03897v2 doi:10.1007/s00180-018-0791-1
Examples
library(GenomicRanges)
library(EnsDb.Hsapiens.v86)
# load data
data('decorateData')
# load gene locations
ensdb = EnsDb.Hsapiens.v86
# Evaluate hierarchical clustering
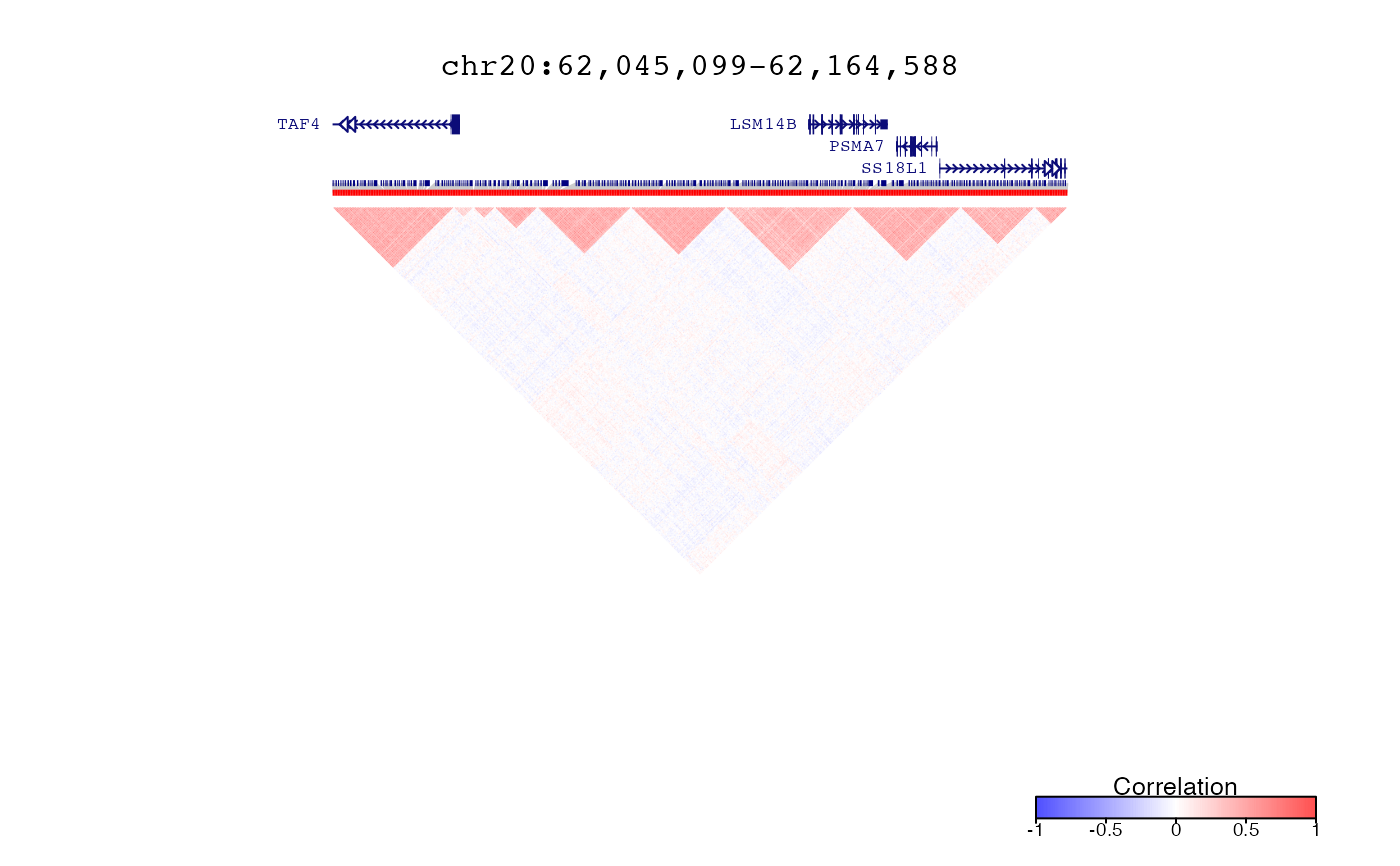
treeList = runOrderedClusteringGenome( simData, simLocation )
#>
Evaluating:chr20
#>
# Choose cutoffs and return clusters
treeListClusters = createClusters( treeList )
#> Method:capushe
# Plot correlations and clusters in region defined by query
query = range(simLocation)
plotDecorate( ensdb, treeList, treeListClusters, simLocation, query)