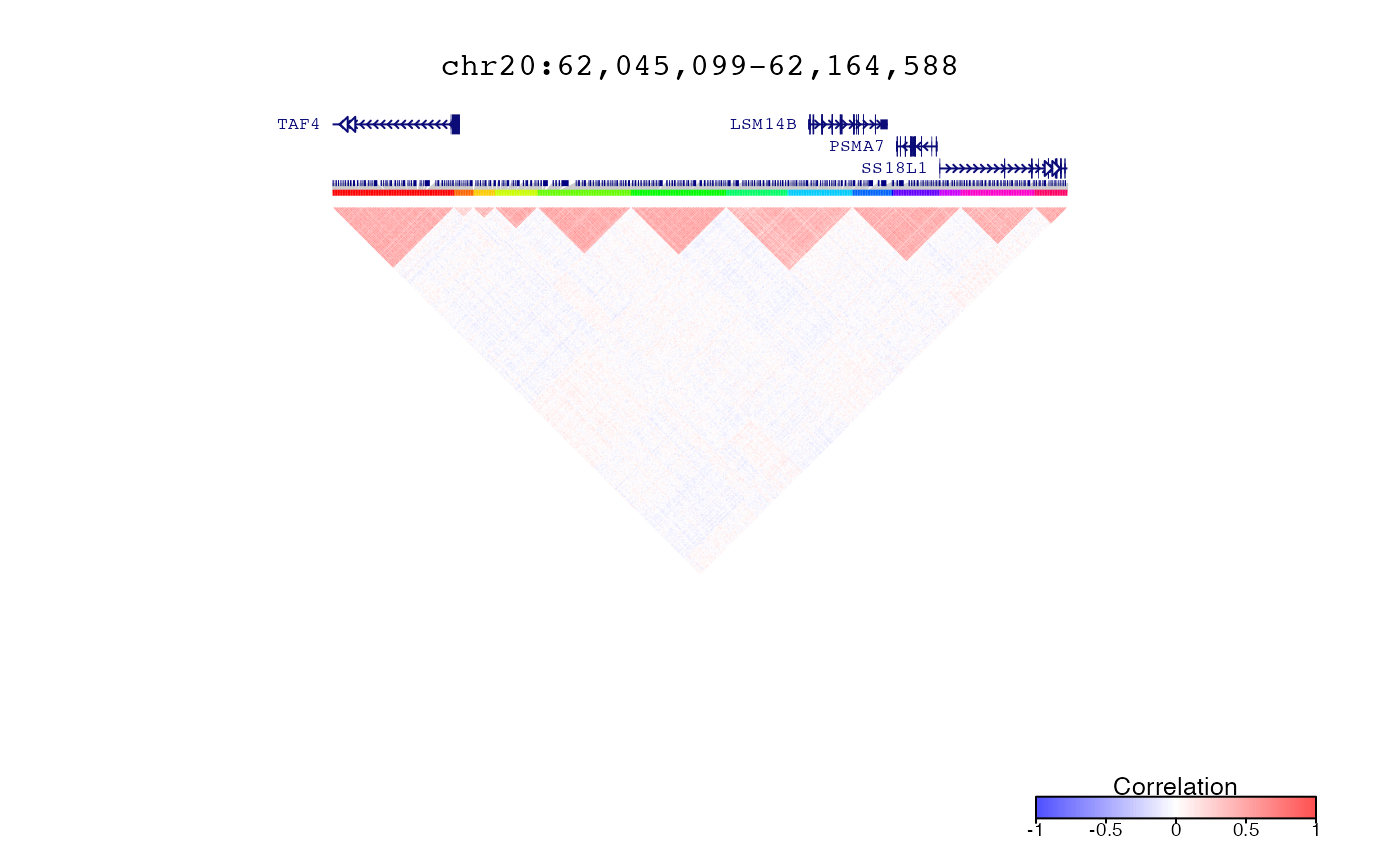
Plot decorate analysis for clusters and correlations
Usage
plotDecorate(
ensdb,
treeList,
treeListClusters,
featurePositions,
query,
data,
cols,
showGenes = TRUE,
splice_variants = FALSE,
non_coding = FALSE,
absCorr = FALSE,
method.corr = c("pearson", "kendall", "spearman")
)Arguments
- ensdb
ENSEMBL database object like EnsDb.Hsapiens.v86
- treeList
hierarchical clustering of each chromosome from runOrderedClusteringGenome()
- treeListClusters
assign regions to clusters after cutting tree with createClusters()
- featurePositions
GRanges object storing location of each feature
- query
GRanges object indiecating region to plot
- data
data to compute correlations stratified by testVariable
- cols
array of color values
- showGenes
plot genes
- splice_variants
if TRUE, show multiple transcripts from the same gene
- non_coding
if TRUE, also show non-coding genes
- absCorr
show absolute correlations
- method.corr
if data is specified, compute correlation using: "pearson", "kendall", "spearman"
Examples
library(GenomicRanges)
library(EnsDb.Hsapiens.v86)
# load data
data('decorateData')
# load gene locations
ensdb = EnsDb.Hsapiens.v86
# Evaluate hierarchical clsutering
treeList = runOrderedClusteringGenome( simData, simLocation )
#>
Evaluating:chr20
#>
# Choose cutoffs and return clusters
treeListClusters = createClusters( treeList, method='meanClusterSize', meanClusterSize=30)
#> Method:meanClusterSize
# Plot correlations and clusters in region defined by query
query = range(simLocation)
plotDecorate( ensdb, treeList, treeListClusters, simLocation, query)