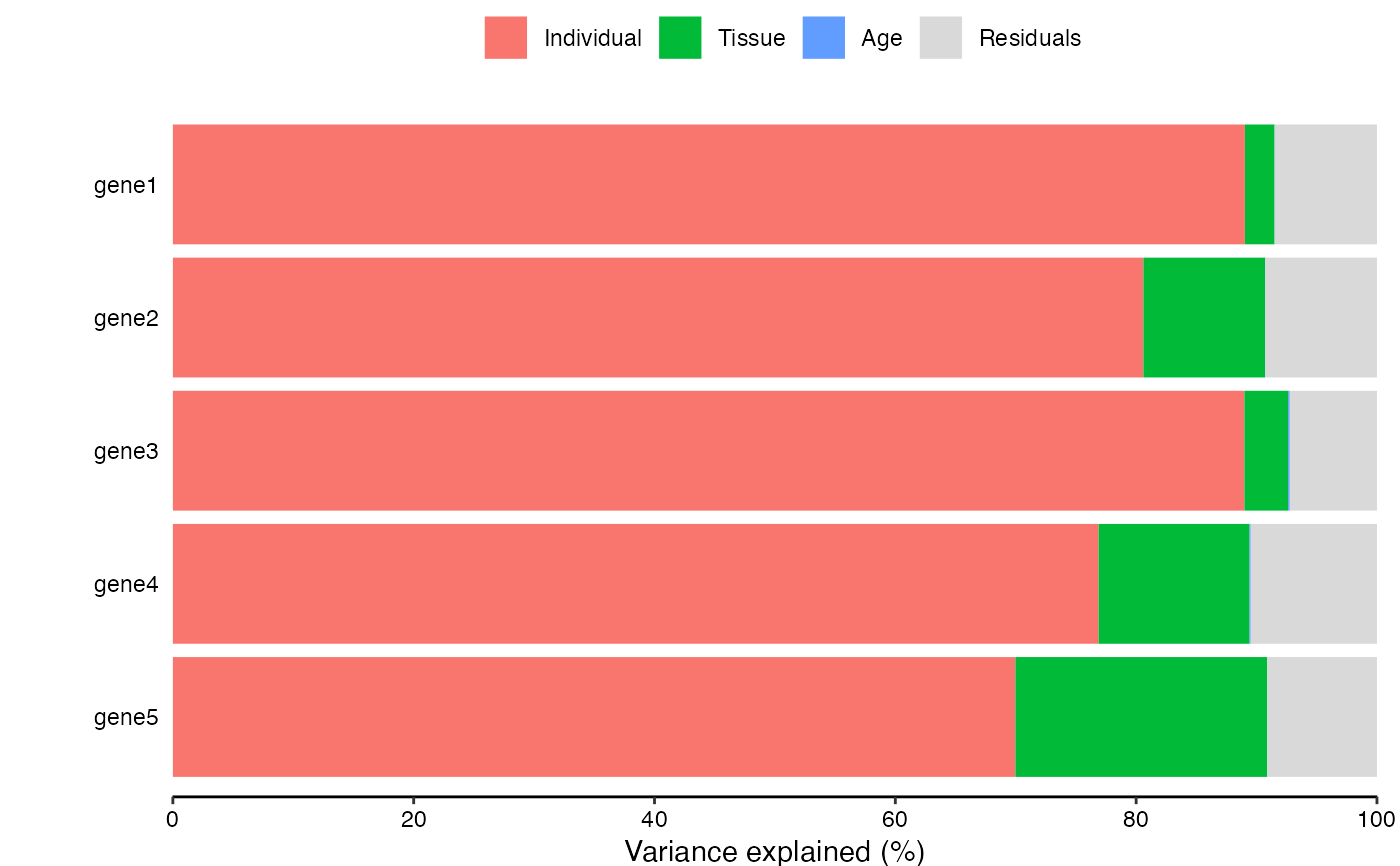
Bar plot of fractions for a subset of genes
Usage
plotPercentBars(
x,
col = c(ggColorHue(ncol(x) - 1), "grey85"),
genes = rownames(x),
width = NULL,
...
)
# S4 method for class 'matrix'
plotPercentBars(
x,
col = c(ggColorHue(ncol(x) - 1), "grey85"),
genes = rownames(x),
width = NULL,
...
)
# S4 method for class 'data.frame'
plotPercentBars(
x,
col = c(ggColorHue(ncol(x) - 1), "grey85"),
genes = rownames(x),
width = NULL,
...
)
# S4 method for class 'varPartResults'
plotPercentBars(
x,
col = c(ggColorHue(ncol(x) - 1), "grey85"),
genes = rownames(x),
width = NULL,
...
)Examples
# library(variancePartition)
library(BiocParallel)
# load simulated data:
# geneExpr: matrix of gene expression values
# info: information/metadata about each sample
data(varPartData)
# Specify variables to consider
form <- ~ Age + (1 | Individual) + (1 | Tissue)
# Fit model
varPart <- fitExtractVarPartModel(geneExpr, form, info)
# Bar plot for a subset of genes showing variance fractions
plotPercentBars(varPart[1:5, ])
#> Warning: Ignoring empty aesthetic: `width`.
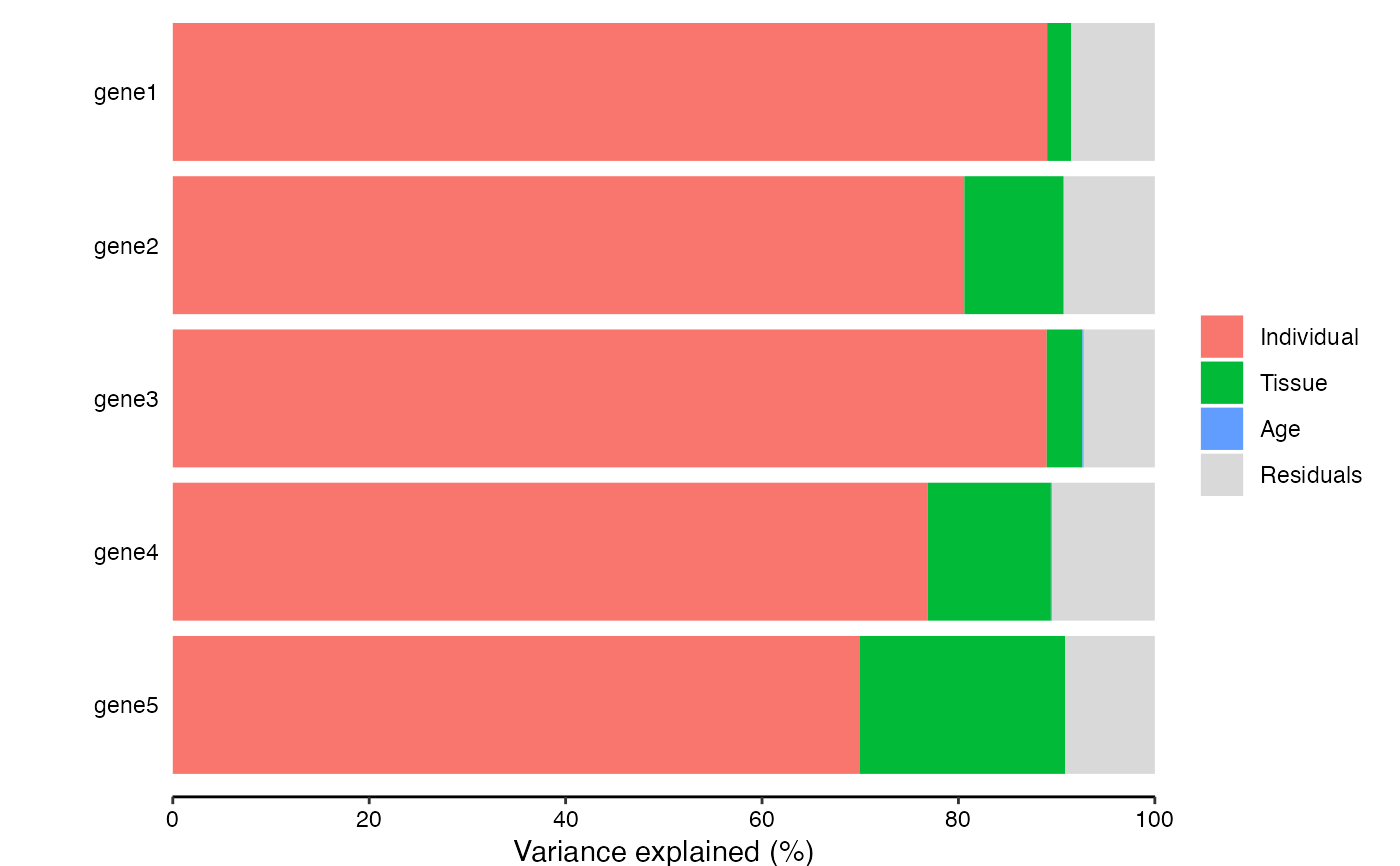 # Move the legend to the top
plotPercentBars(varPart[1:5, ]) + theme(legend.position = "top")
#> Warning: Ignoring empty aesthetic: `width`.
# Move the legend to the top
plotPercentBars(varPart[1:5, ]) + theme(legend.position = "top")
#> Warning: Ignoring empty aesthetic: `width`.