Plot correlation structure of a gene based on random effects
Usage
plotCorrStructure(
fit,
varNames = names(coef(fit)),
reorder = TRUE,
pal = colorRampPalette(c("white", "red", "darkred")),
hclust.method = "complete"
)Arguments
- fit
linear mixed model fit of a gene produced by lmer() or fitVarPartModel()
- varNames
variables in the metadata for which the correlation structure should be shown. Variables must be random effects
- reorder
how to reorder the rows/columns of the correlation matrix. reorder=FALSE gives no reorder. reorder=TRUE reorders based on hclust. reorder can also be an array of indices to reorder the samples manually
- pal
color palette
- hclust.method
clustering methods for hclust
Examples
# load library
# library(variancePartition)
library(BiocParallel)
# load simulated data:
data(varPartData)
# specify formula
form <- ~ Age + (1 | Individual) + (1 | Tissue)
# fit and return linear mixed models for each gene
fitList <- fitVarPartModel(geneExpr[1:10, ], form, info)
# Focus on the first gene
fit <- fitList[[1]]
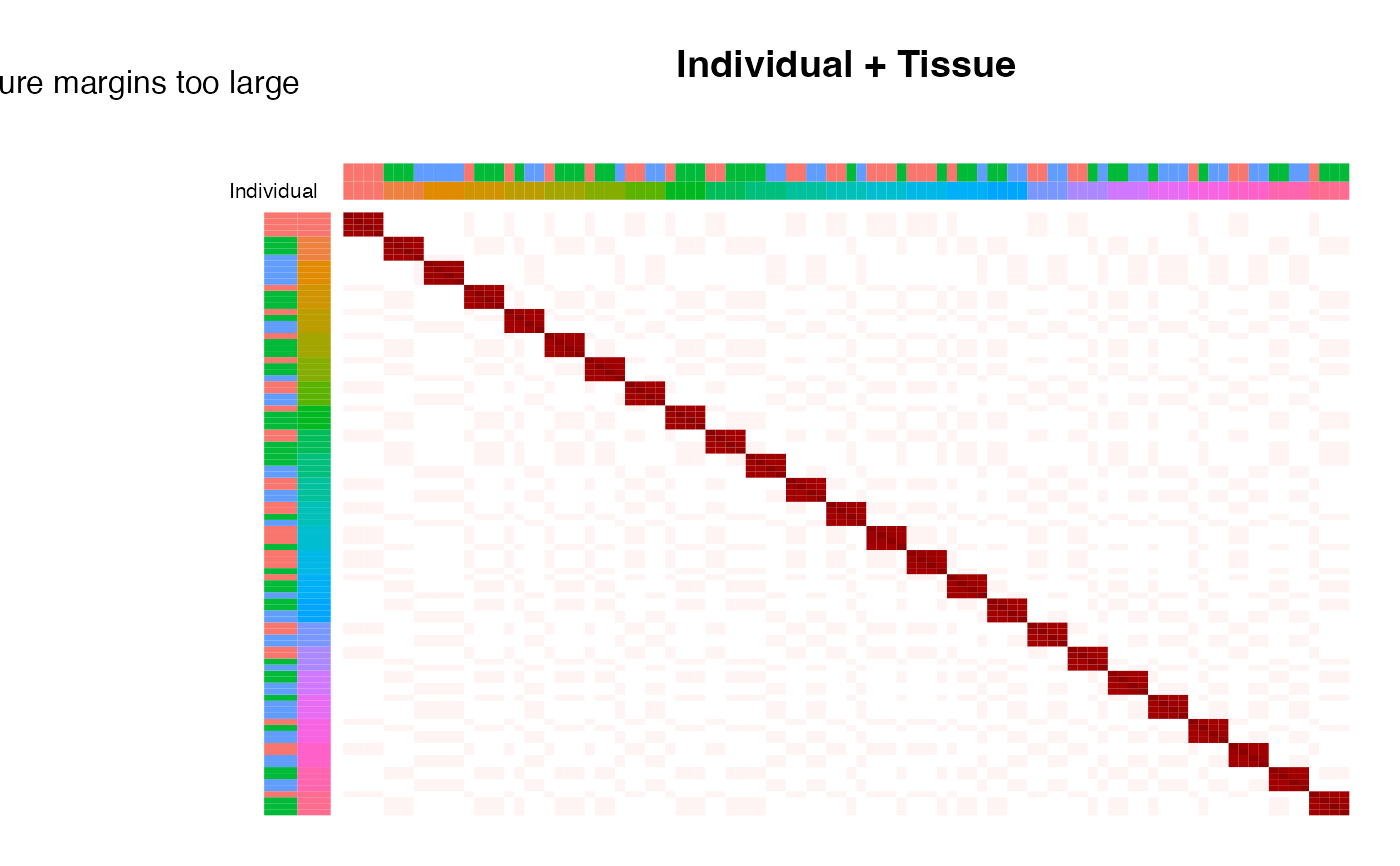
# plot correlation sturcture based on Individual, reordering samples with hclust
plotCorrStructure(fit, "Individual")
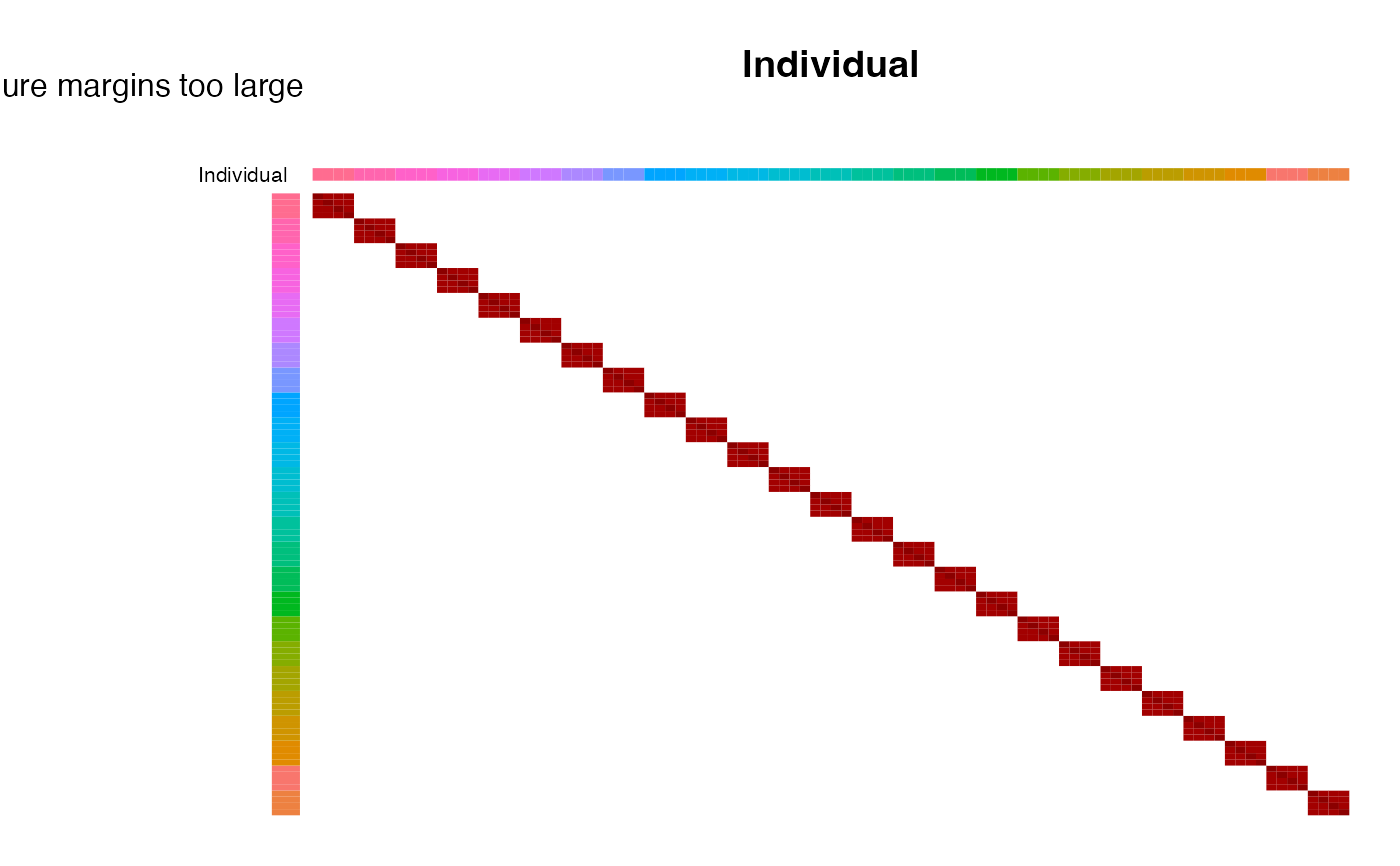 # don't reorder
plotCorrStructure(fit, "Individual", reorder = FALSE)
# don't reorder
plotCorrStructure(fit, "Individual", reorder = FALSE)
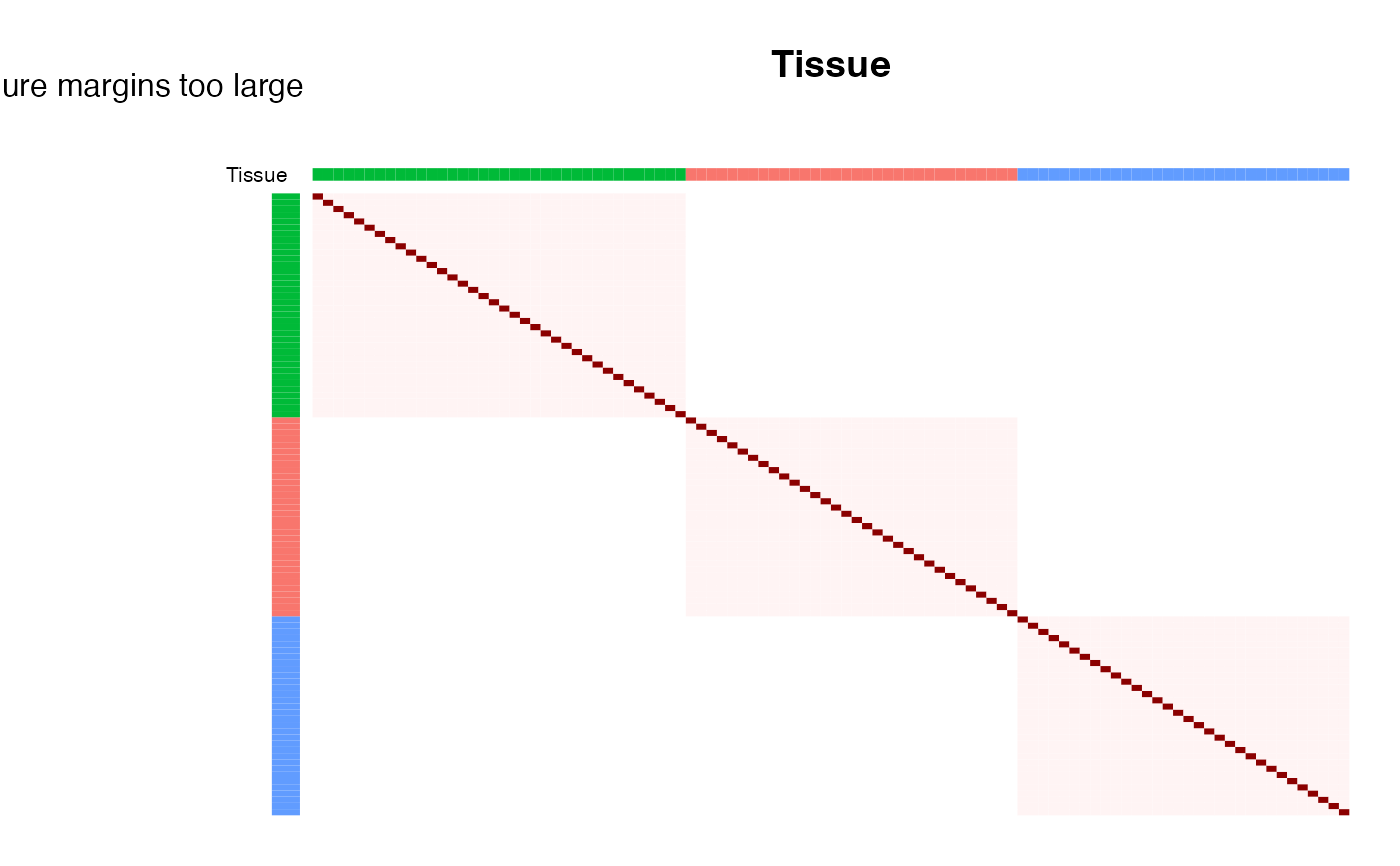
 # plot correlation sturcture based on Tissue, reordering samples with hclust
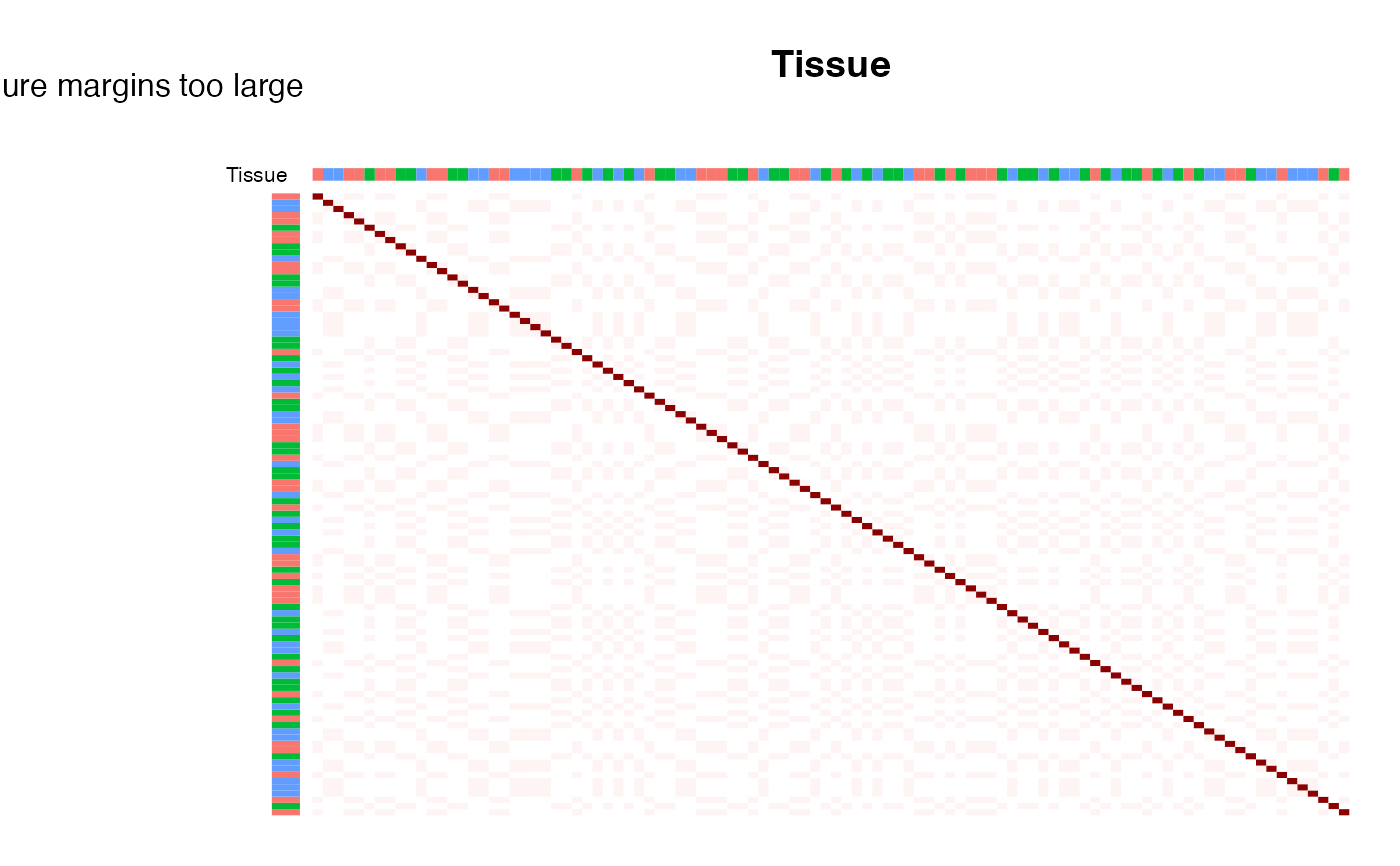
plotCorrStructure(fit, "Tissue")
# plot correlation sturcture based on Tissue, reordering samples with hclust
plotCorrStructure(fit, "Tissue")
 # don't reorder
plotCorrStructure(fit, "Tissue", FALSE)
# don't reorder
plotCorrStructure(fit, "Tissue", FALSE)
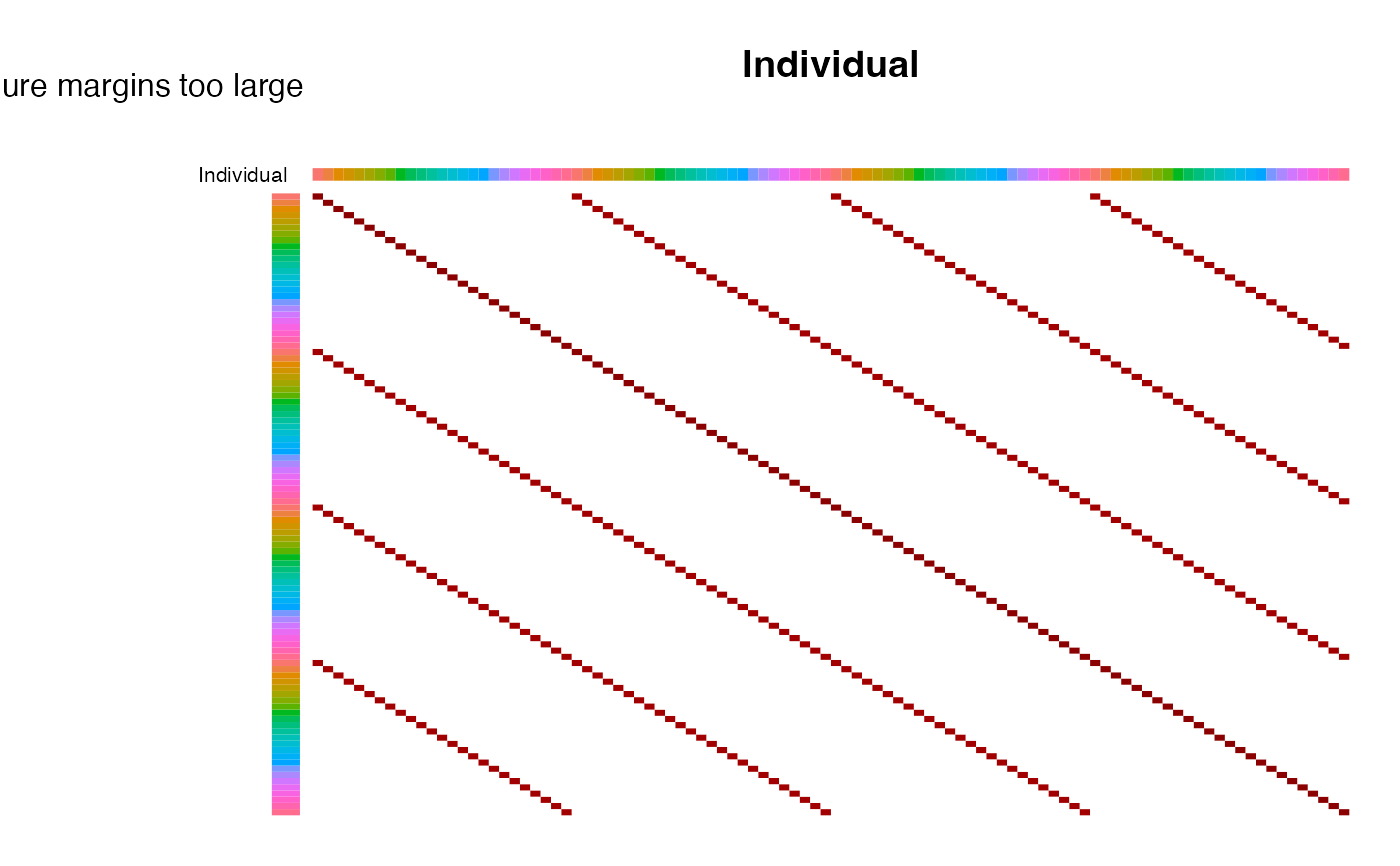
 # plot correlation structure based on all random effects
# reorder manually by Tissue and Individual
idx <- order(info$Tissue, info$Individual)
plotCorrStructure(fit, reorder = idx)
# plot correlation structure based on all random effects
# reorder manually by Tissue and Individual
idx <- order(info$Tissue, info$Individual)
plotCorrStructure(fit, reorder = idx)
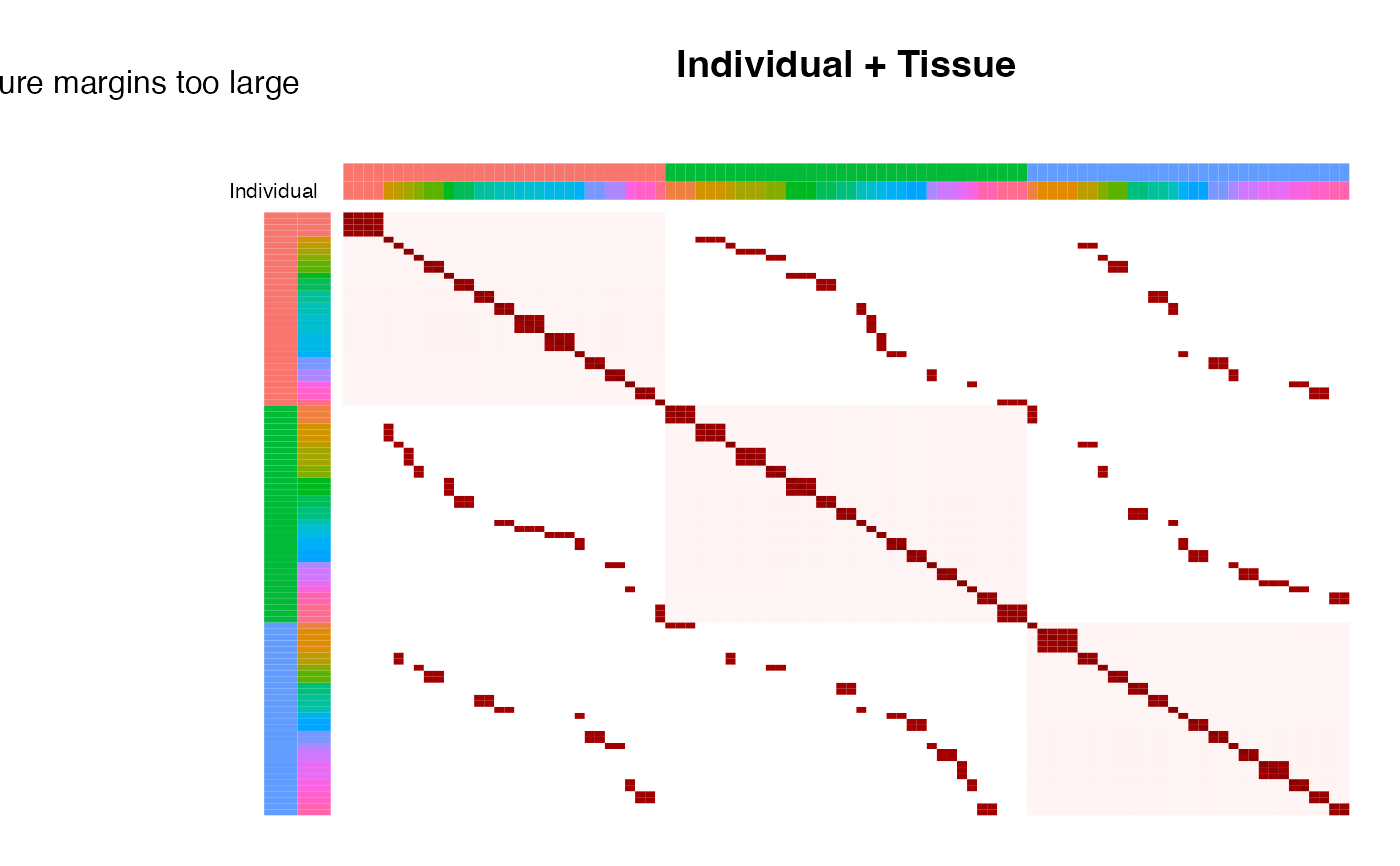 # plot correlation structure based on all random effects
# reorder manually by Individual, then Tissue
idx <- order(info$Individual, info$Tissue)
plotCorrStructure(fit, reorder = idx)
# plot correlation structure based on all random effects
# reorder manually by Individual, then Tissue
idx <- order(info$Individual, info$Tissue)
plotCorrStructure(fit, reorder = idx)