Approximating logistic coefficients
A simulation analysis
Developed by Gabriel Hoffman
Run on 2025-07-28 10:48:36.933734
Source:vignettes/beta_approx.Rmd
beta_approx.RmdGiven a z-statistic, we want to obtain the coefficient value from a linear regression. Adapting the approach from Zhu, et al (2016, Methods eqn 6), we estimate the coefficient as \beta_{linear} = z * sd_y / (sd_x*sqrt(n + z^2)), where sd_x is the standard deviation of the covariate and sd_y is the standard error of the response. For a model with no covariates, this transformation gives the exact coefficient estimate. With covariates, it is approximate.
The coeffient estimate from linear regression can be converted to the
logistic scale using the approach of Pirinen, et al. (2013)
according to \beta_{logistic} =
\beta_{linear} / (\phi(1-\phi)), where \phi is the case ratio in the logistic
regression. This approximates the coefficient as if the model had been
fit with logistic regression. This is implemented in
coef_from_z().
We see here that using the z-statistic to approximate the coefficient from logistic regression model is most accurate for large sample size, balanced class ratio (i.e. \phi near 0.5), and small \beta.
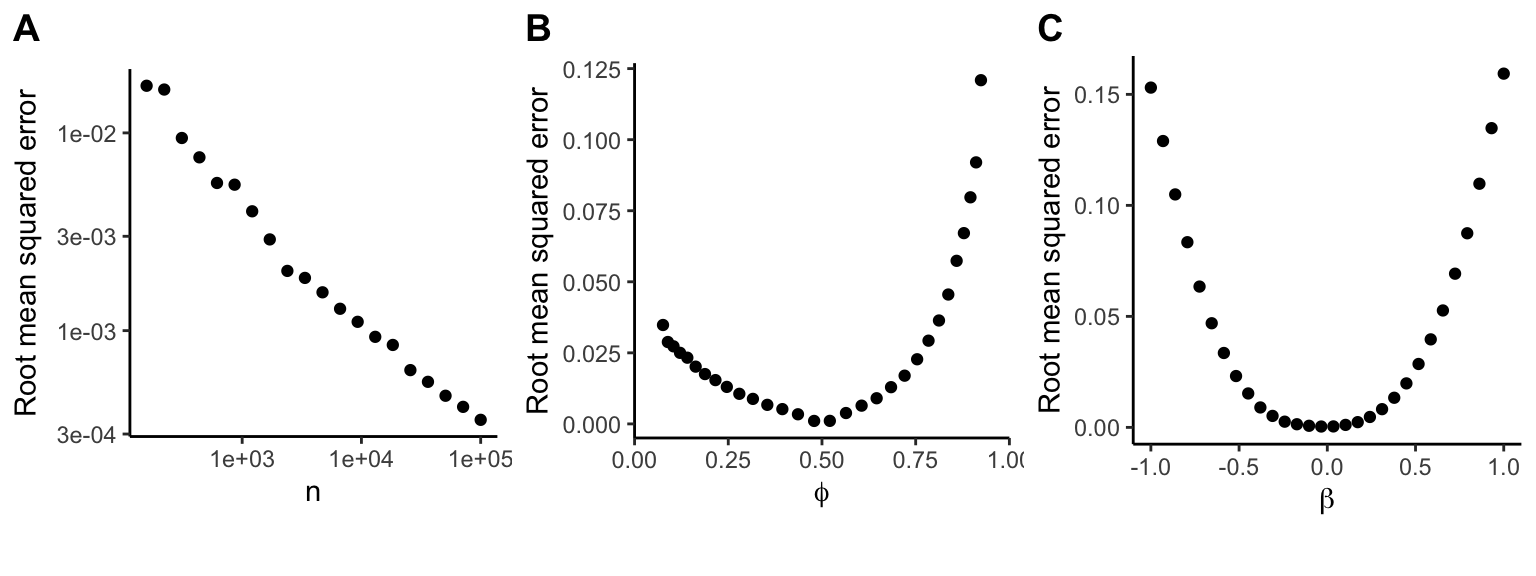
Figure 1. Accuracy of approximating \beta from z-statistics. A) Sample size is varied with \beta=0, \phi=0.5 B) Case ratio is varied with \beta=0, n=10,000 C) \beta is varied with \phi=0.5, n=10,000
Session Info
## R version 4.4.2 (2024-10-31)
## Platform: aarch64-apple-darwin23.6.0
## Running under: macOS Sonoma 14.7.1
##
## Matrix products: default
## BLAS: /Users/gabrielhoffman/prog/R-4.4.2/lib/libRblas.dylib
## LAPACK: /opt/homebrew/Cellar/r/4.5.1/lib/R/lib/libRlapack.dylib; LAPACK version 3.12.1
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## time zone: America/New_York
## tzcode source: internal
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] cowplot_1.1.3 lubridate_1.9.4 forcats_1.0.0 stringr_1.5.1
## [5] dplyr_1.1.4 purrr_1.0.4 readr_2.1.5 tidyr_1.3.1
## [9] tibble_3.3.0 ggplot2_3.5.2 tidyverse_2.0.0 imputez_1.2.3
##
## loaded via a namespace (and not attached):
## [1] Rdpack_2.6.4 rlang_1.1.6
## [3] magrittr_2.0.3 matrixStats_1.5.0
## [5] compiler_4.4.2 systemfonts_1.2.3
## [7] vctrs_0.6.5 pkgconfig_2.0.3
## [9] crayon_1.5.3 fastmap_1.2.0
## [11] XVector_0.46.0 labeling_0.4.3
## [13] CholWishart_1.1.4 rmarkdown_2.29
## [15] tzdb_0.5.0 pracma_2.4.4
## [17] UCSC.utils_1.2.0 ragg_1.4.0
## [19] xfun_0.52 Rfast_2.1.5.1
## [21] zlibbioc_1.52.0 cachem_1.1.0
## [23] beachmat_2.22.0 GenomeInfoDb_1.42.3
## [25] jsonlite_2.0.0 progress_1.2.3
## [27] rhdf5filters_1.18.1 DelayedArray_0.32.0
## [29] Rhdf5lib_1.28.0 irlba_2.3.5.1
## [31] parallel_4.4.2 prettyunits_1.2.0
## [33] R6_2.6.1 bslib_0.9.0
## [35] stringi_1.8.7 RColorBrewer_1.1-3
## [37] GenomicRanges_1.58.0 jquerylib_0.1.4
## [39] Rcpp_1.0.14 SummarizedExperiment_1.36.0
## [41] iterators_1.0.14 knitr_1.50
## [43] CovTools_0.5.4 base64enc_0.1-3
## [45] IRanges_2.40.1 timechange_0.3.0
## [47] Matrix_1.7-3 igraph_2.1.4
## [49] tidyselect_1.2.1 abind_1.4-8
## [51] yaml_2.3.10 doParallel_1.0.17
## [53] codetools_0.2-20 minpack.lm_1.2-4
## [55] lattice_0.22-7 withr_3.0.2
## [57] Biobase_2.66.0 evaluate_1.0.4
## [59] desc_1.4.3 geigen_2.3
## [61] RcppParallel_5.1.10 pillar_1.10.2
## [63] MatrixGenerics_1.18.1 ShrinkCovMat_1.4.0
## [65] foreach_1.5.2 SHT_0.1.9
## [67] stats4_4.4.2 generics_0.1.4
## [69] S4Vectors_0.44.0 hms_1.1.3
## [71] scales_1.4.0 flare_1.7.0.2
## [73] glue_1.8.0 GenomicDataStream_0.0.60
## [75] scatterplot3d_0.3-44 tools_4.4.2
## [77] beachmat.hdf5_1.4.0 fs_1.6.6
## [79] mvtnorm_1.3-3 rgl_1.3.18
## [81] rhdf5_2.50.2 grid_4.4.2
## [83] rbibutils_2.3 GenomeInfoDbData_1.2.13
## [85] HDF5Array_1.34.0 cli_3.6.5
## [87] zigg_0.0.2 textshaping_1.0.1
## [89] expm_1.0-0 S4Arrays_1.6.0
## [91] corpcor_1.6.10 gtable_0.3.6
## [93] shapes_1.2.7 sass_0.4.10
## [95] digest_0.6.37 BiocGenerics_0.52.0
## [97] SparseArray_1.6.2 htmlwidgets_1.6.4
## [99] farver_2.1.2 htmltools_0.5.8.1
## [101] pkgdown_2.1.3 decorrelate_0.1.6.3
## [103] lifecycle_1.0.4 httr_1.4.7
## [105] MASS_7.3-65