Software

crumblr
Fast, flexible analysis of differences in cellular composition with crumblr
dreamlet
Efficient differential expression analysis of large-scale single cell transcriptomics data
decorate
decorate: Differential Epigenetic Coregeulation Test
dream
dream: Powerful differential expression analysis for repeated measures designs
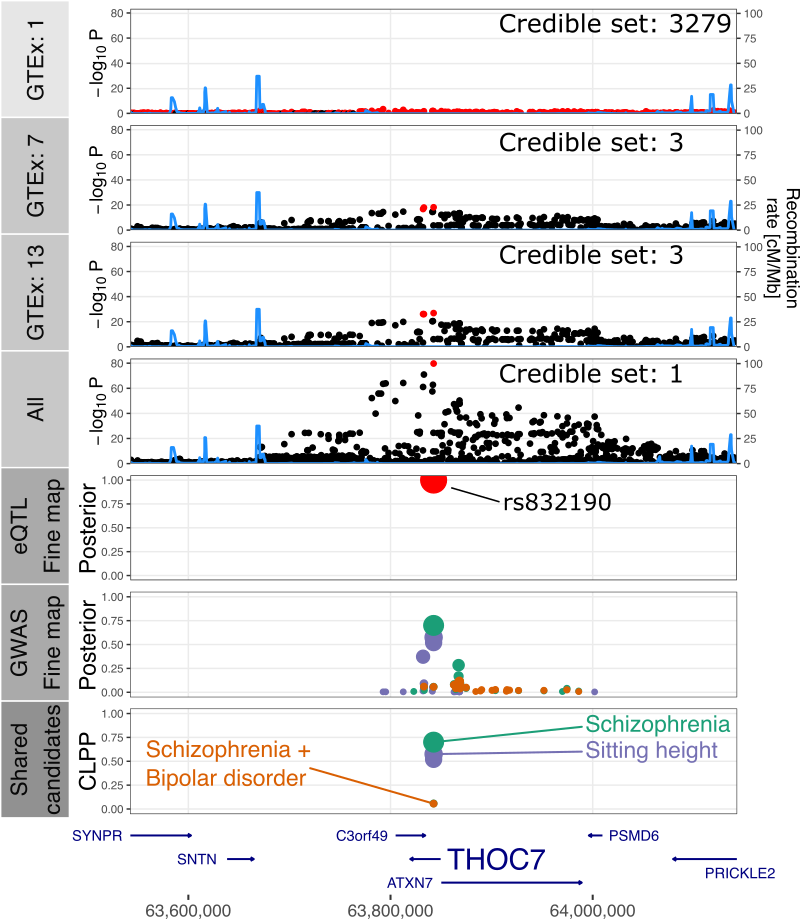
Brain eQTL meta-analysis
Joint fine-mapping of large-scale post-mortem brain resource with many public GWAS datasets

CommonMind Consortium provides transcriptomic and epigenomic data for Schizophrenia and Bipolar Disorder
Public resource of functional genomic data from the dorsolateral prefrontal cortex (DLPFC; Brodmann areas 9 and 46): RNA-seq and SNP genotypes on 980 individuals, and ATAC-seq on 269 individuals from 4 separate brain banks.
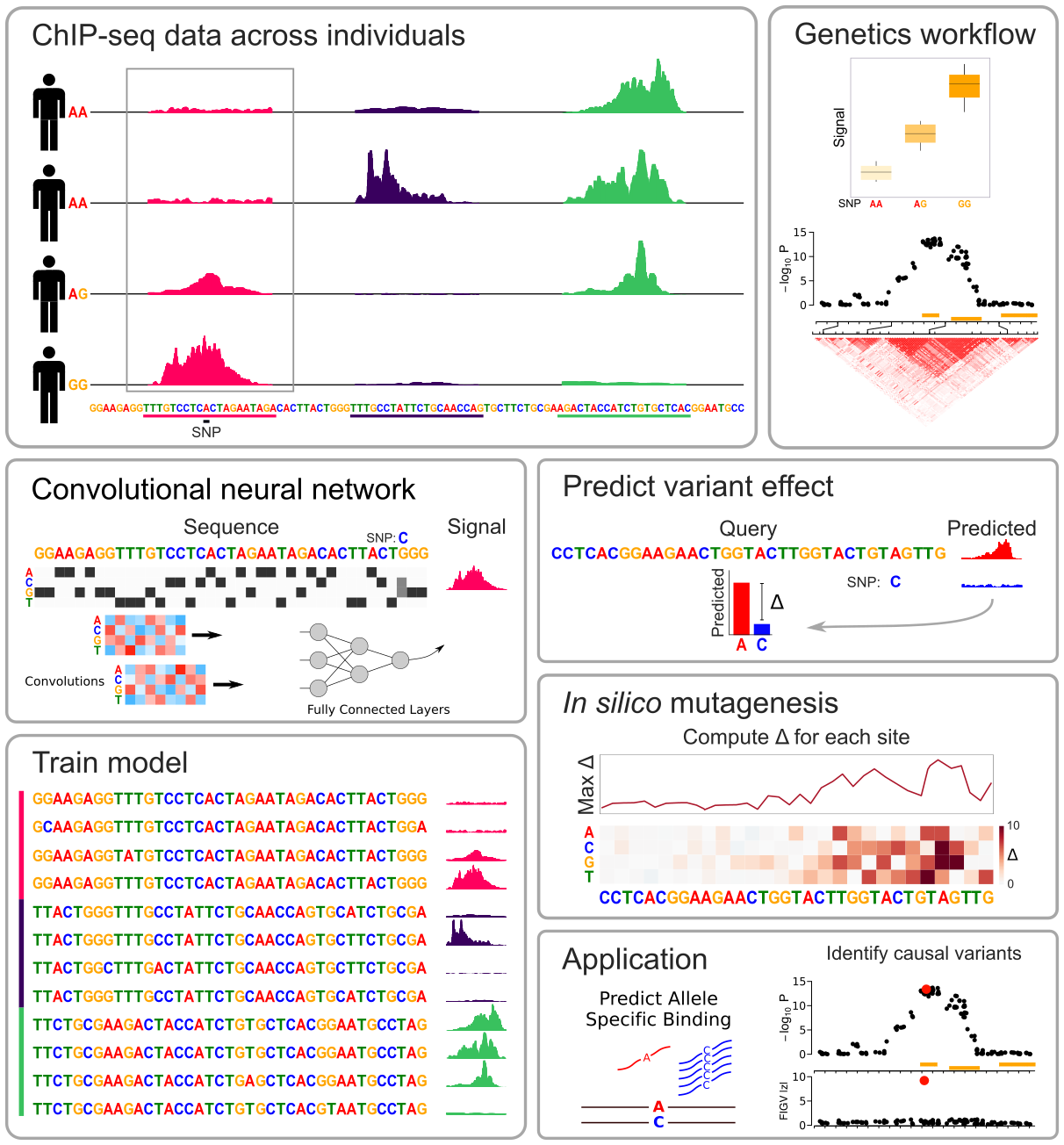
DeepFIGV
Functional Interpretation of Genetic Variants Using Deep Learning Predicts Impact on Epigenome.
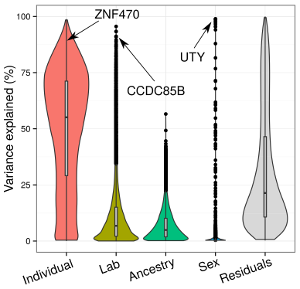
variancePartition
Interpreting drivers of variation in complex gene expression studies with linear mixed models
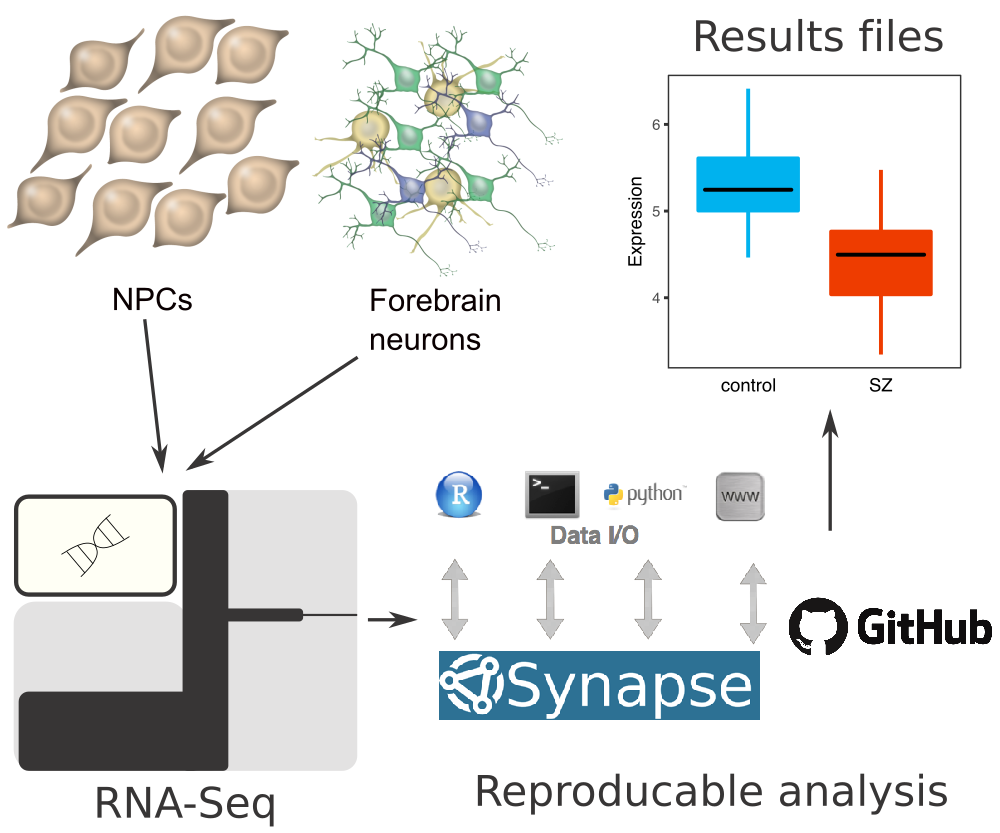
Data and code resources for an hiPSC model
RNA-seq data from hiPSC-derived neural progenitor cells and neurons from controls and patients with childhood onset schizophrenia
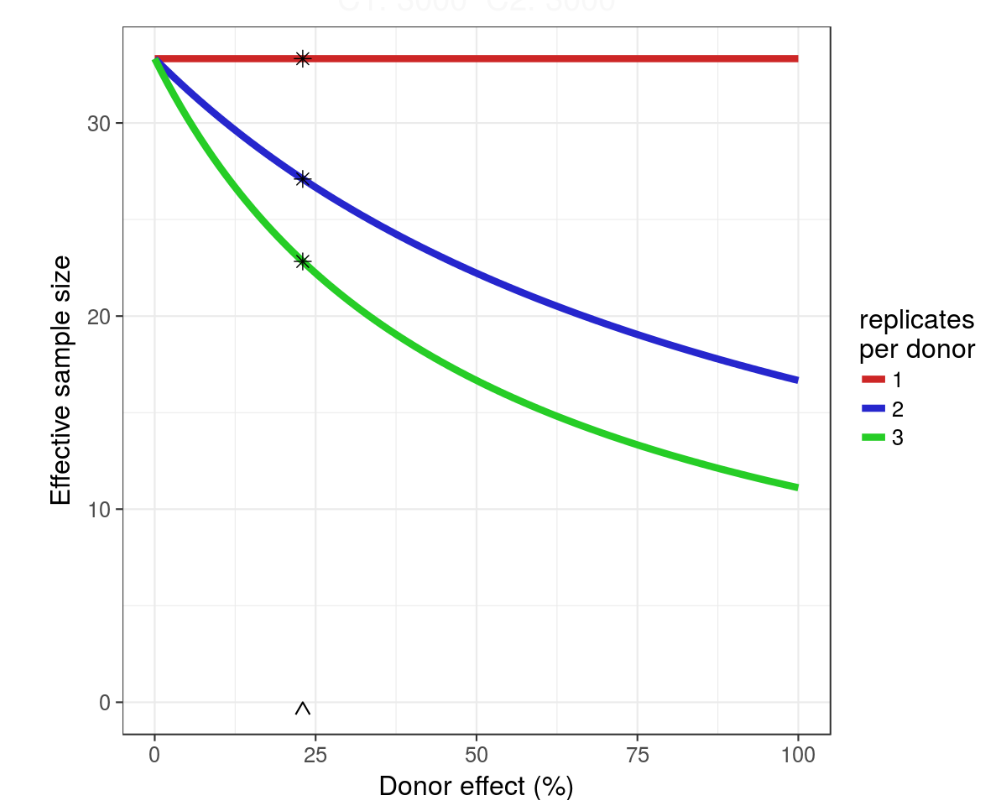
Design hiPSC experiments
Design powerful transcriptome experiments given cost constraints
Referance map of human brain epigenome
Data from ChIP-seq for H3K4me3 (promoters) and H3K27ac (enhancers and promoters) from 2 brain regions from 17 individuals

Epigenomics
PsychENCODE

lrgpr
Interactive linear mixed model analysis of genome-wide association studies with composite hypothesis testing and regression diagnostics in R