 |
GenomicDataStream
A scalable interface between data and analysis
|
 |
GenomicDataStream
A scalable interface between data and analysis
|
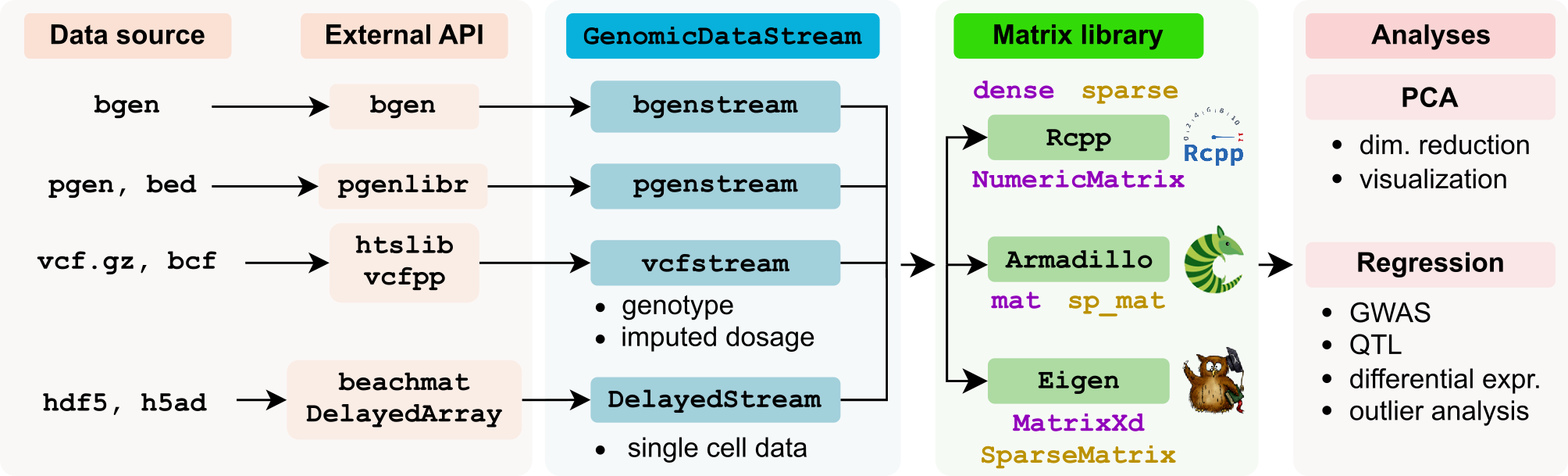
GenomicDataStream provides interfaces at both the C++ and R levels. The C++ interface prioritizes efficiency, while the R interface wraps the C++ backend for non-technical users.
| Package | Ref | Role |
|---|---|---|
| vcfppR | Bioinformatics | C++ API for htslib |
| htslib | GigaScience | C API for VCF/BCF files |
| pgenlibr | GigaScience | R/C++ API for plink files |
| beatchmat | PLoS Comp Biol | C++ API for access data owned by R |
| Rcpp | J Stat Software | API for R/C++ integration |
| RcppEigen | J Stat Software | API for Rcpp access to Eigen matrix library |
| RcppArmadillo | J Stat Software | API for Rcpp access to Armadillo matrix library |
| Eigen | C++ library for linear algebra with advanced features | |
| Armadillo | J Open Src Soft | User-friendly C++ library for linear algebra |
| RcppParallel | oneAPI Threading Building Blocks for parallel analysis |
-D DISABLE_DELAYED_STREAM
Omit DelayedStream class, remove dependence on Rcpp and beachmat
-D DISABLE_EIGEN
Omit support for Eigen matrix library, and remove dependence on RcppEigen and Eigen
-D DISABLE_RCPP
Omit support for Rcpp matrix library, and remove dependence on Rcpp
-D DISABLE_PLINK
Omit support for PLINK files (PGEN, BED), and remove dependence on pgenlibr
 Developed by Gabriel Hoffman at Center for Disease Neurogenomics at the Icahn School of Medicine at Mount Sinai.
Developed by Gabriel Hoffman at Center for Disease Neurogenomics at the Icahn School of Medicine at Mount Sinai.